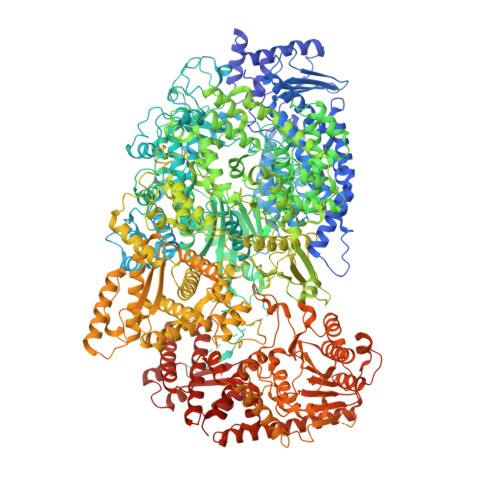
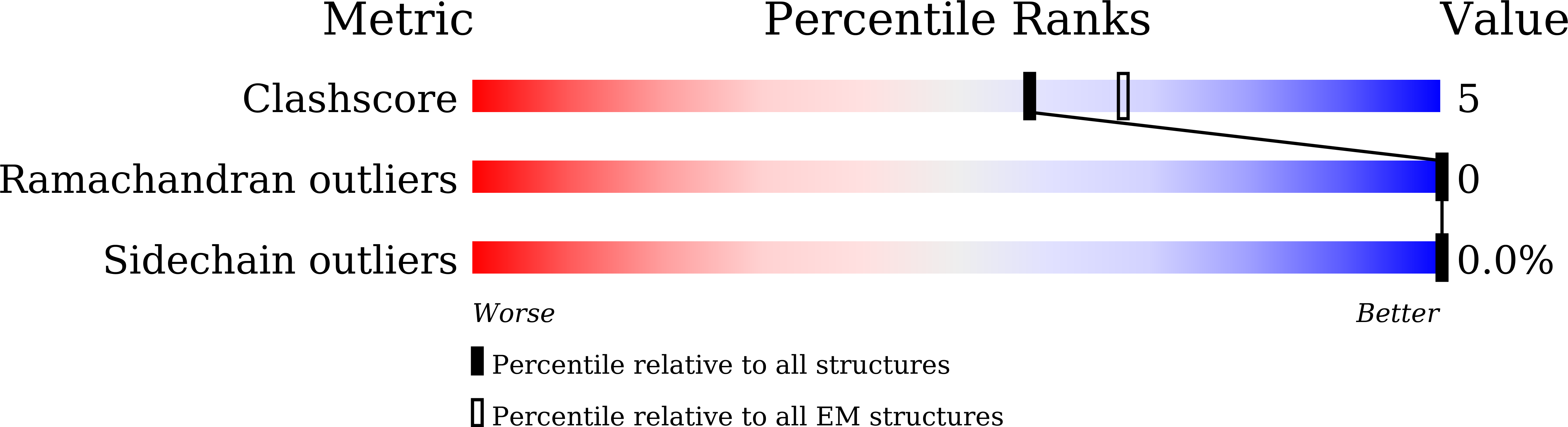Structural basis for dimerization of a paramyxovirus polymerase complex.
Xie, J., Ouizougun-Oubari, M., Wang, L., Zhai, G., Wu, D., Lin, Z., Wang, M., Ludeke, B., Yan, X., Nilsson, T., Gao, L., Huang, X., Fearns, R., Chen, S.(2024) Nat Commun 15: 3163-3163
- PubMed: 38605025
- DOI: https://doi.org/10.1038/s41467-024-47470-7
- Primary Citation of Related Structures:
8KDB, 8KDC - PubMed Abstract:
The transcription and replication processes of non-segmented, negative-strand RNA viruses (nsNSVs) are catalyzed by a multi-functional polymerase complex composed of the large protein (L) and a cofactor protein, such as phosphoprotein (P). Previous studies have shown that the nsNSV polymerase can adopt a dimeric form, however, the structure of the dimer and its function are poorly understood. Here we determine a 2.7 Å cryo-EM structure of human parainfluenza virus type 3 (hPIV3) L-P complex with the connector domain (CD') of a second L built, while reconstruction of the rest of the second L-P obtains a low-resolution map of the ring-like L core region. This study reveals detailed atomic features of nsNSV polymerase active site and distinct conformation of hPIV3 L with a unique β-strand latch. Furthermore, we report the structural basis of L-L dimerization, with CD' located at the putative template entry of the adjoining L. Disruption of the L-L interface causes a defect in RNA replication that can be overcome by complementation, demonstrating that L dimerization is necessary for hPIV3 genome replication. These findings provide further insight into how nsNSV polymerases perform their functions, and suggest a new avenue for rational drug design.
Organizational Affiliation:
Roche Pharma Research and Early Development, Lead Discovery, Roche Innovation Center Shanghai, 201203, Shanghai, China.