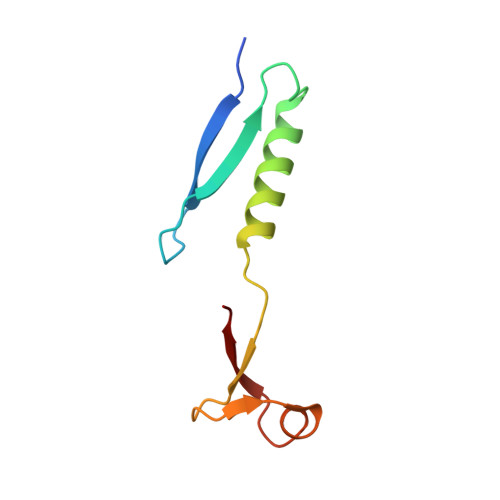
Domain swap in the C-terminal ubiquitin-like domain of human doublecortin.
Rufer, A.C., Kusznir, E., Burger, D., Stihle, M., Ruf, A., Rudolph, M.G.(2018) Acta Crystallogr D Struct Biol 74: 450-462
- PubMed: 29717716
- DOI: https://doi.org/10.1107/S2059798318004813
- Primary Citation of Related Structures:
6FNZ - PubMed Abstract:
Doublecortin, a microtubule-associated protein that is only produced during neurogenesis, cooperatively binds to microtubules and stimulates microtubule polymerization and cross-linking by unknown mechanisms. A domain swap is observed in the crystal structure of the C-terminal domain of doublecortin. As determined by analytical ultracentrifugation, an open conformation is also present in solution. At higher concentrations, higher-order oligomers of the domain are formed. The domain swap and additional interfaces observed in the crystal lattice can explain the formation of doublecortin tetramers or multimers, in line with the analytical ultracentrifugation data. Taken together, the domain swap offers a mechanism for the observed cooperative binding of doublecortin to microtubules. Doublecortin-induced cross-linking of microtubules can be explained by the same mechanism. The effect of several mutations leading to lissencephaly and double-cortex syndrome can be traced to the domain swap and the proposed self-association of doublecortin.
Organizational Affiliation:
pRED, Therapeutic Modalities, F. Hoffmann-La Roche, 4070 Basel, Switzerland.