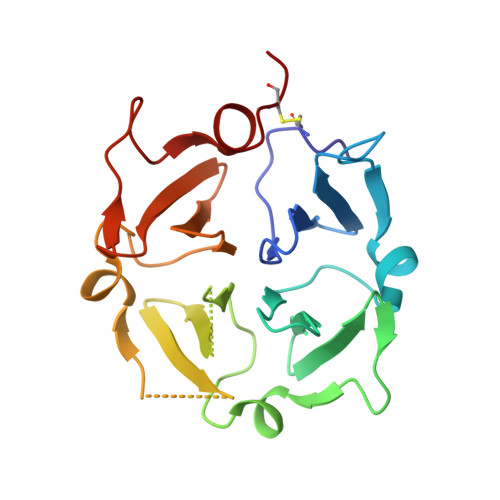
Structure of human Vitronectin C-terminal domain and interaction withYersinia pestisouter membrane protein Ail.
Shin, K., Lechtenberg, B.C., Fujimoto, L.M., Yao, Y., Bartra, S.S., Plano, G.V., Marassi, F.M.(2019) Sci Adv 5: eaax5068-eaax5068
- PubMed: 31535027
- DOI: https://doi.org/10.1126/sciadv.aax5068
- Primary Citation of Related Structures:
6O5E - PubMed Abstract:
Vitronectin (Vn) is a major component of blood that controls many processes central to human biology. It is a drug target and a key factor in cell and tissue engineering applications, but despite long-standing efforts, little is known about the molecular basis for its functions. Here, we define the domain organization of Vn, report the crystal structure of its carboxyl-terminal domain, and show that it harbors the binding site for the Yersinia pestis outer membrane protein Ail, which recruits Vn to the bacterial cell surface to evade human host defenses. Vn forms a single four-bladed β/α-propeller that serves as a hub for multiple functions. The structure explains key features of native Vn and provides a blueprint for understanding and targeting this essential human protein.
Organizational Affiliation:
Cancer Center, Sanford Burnham Prebys Medical Discovery Institute, 10901 North Torrey Pines Road, La Jolla, CA 92037, USA.