First SAR Study for Overriding NRAS Mutant Driven Acute Myeloid Leukemia.
Cho, H., Shin, I., Ju, E., Choi, S., Hur, W., Kim, H., Hong, E., Kim, N.D., Choi, H.G., Gray, N.S., Sim, T.(2018) J Med Chem 61: 8353-8373
- PubMed: 30153003
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00882
- Primary Citation of Related Structures:
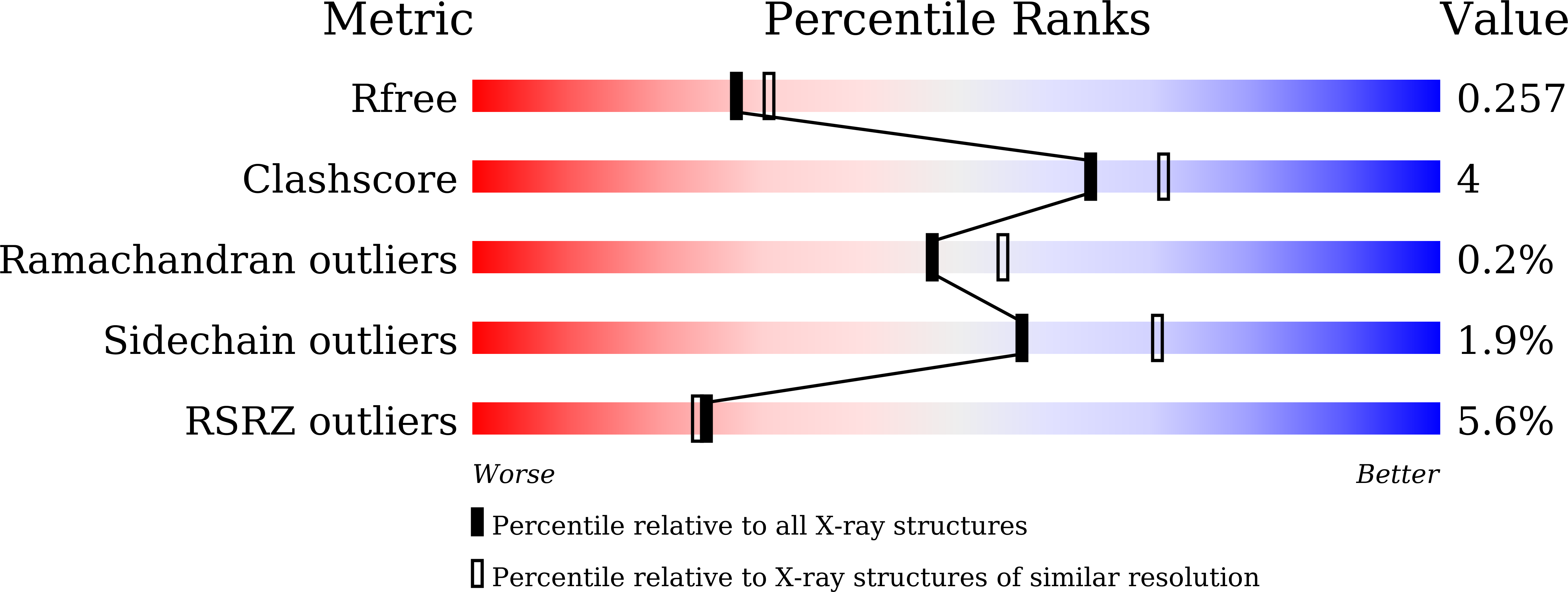
5ZXB - PubMed Abstract:
GNF-7, a multitargeted kinase inhibitor, served as a dual kinase inhibitor of ACK1 and GCK, which provided a novel therapeutic strategy for overriding AML expressing NRAS mutation. This SAR study with GNF-7 derivatives, designed to target NRAS mutant-driven AML, led to identification of the extremely potent inhibitors, 10d, 10g, and 11i, which possess single-digit nanomolar inhibitory activity against both ACK1 and GCK. These substances strongly suppress proliferation of mutant NRAS expressing AML cells via apoptosis and AKT/mTOR signaling blockade. Compound 11i is superior to GNF-7 in terms of kinase inhibitory activity, cellular activity, and differential cytotoxicity. Moreover, 10k possessing a favorable mouse pharmacokinetic profile prolonged life-span of Ba/F3-NRAS-G12D injected mice and significantly delayed tumor growth of OCI-AML3 xenograft model without causing the prominent level of toxicity found with GNF-7. Taken together, this study provides insight into the design of novel ACK1 and GCK dual inhibitors for overriding NRAS mutant-driven AML.
Organizational Affiliation:
KU-KIST Graduate School of Converging Science and Technology , Korea University , 145 Anam-ro, Seongbuk-gu , Seoul 02841 , Republic of Korea.