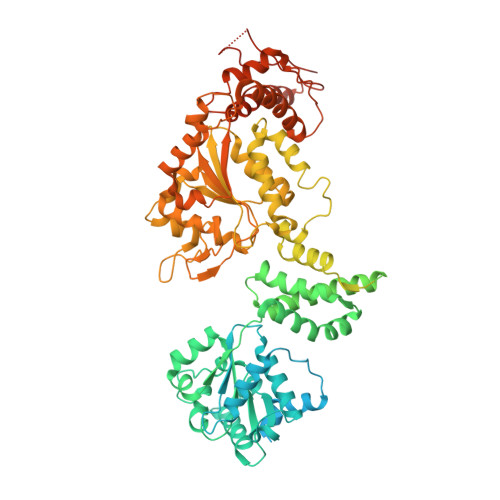
Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Gates, S.N., Yokom, A.L., Lin, J., Jackrel, M.E., Rizo, A.N., Kendsersky, N.M., Buell, C.E., Sweeny, E.A., Mack, K.L., Chuang, E., Torrente, M.P., Su, M., Shorter, J., Southworth, D.R.(2017) Science 357: 273-279
- PubMed: 28619716
- DOI: https://doi.org/10.1126/science.aan1052
- Primary Citation of Related Structures:
5VJH, 5VY8, 5VY9, 5VYA - PubMed Abstract:
Hsp100 polypeptide translocases are conserved members of the AAA+ family (adenosine triphosphatases associated with diverse cellular activities) that maintain proteostasis by unfolding aberrant and toxic proteins for refolding or proteolytic degradation. The Hsp104 disaggregase from Saccharomyces cerevisiae solubilizes stress-induced amorphous aggregates and amyloids. The structural basis for substrate recognition and translocation is unknown. Using a model substrate (casein), we report cryo-electron microscopy structures at near-atomic resolution of Hsp104 in different translocation states. Substrate interactions are mediated by conserved, pore-loop tyrosines that contact an 80-angstrom-long unfolded polypeptide along the axial channel. Two protomers undergo a ratchet-like conformational change that advances pore loop-substrate interactions by two amino acids. These changes are coupled to activation of specific nucleotide hydrolysis sites and, when transmitted around the hexamer, reveal a processive rotary translocation mechanism and substrate-responsive flexibility during Hsp104-catalyzed disaggregation.
Organizational Affiliation:
Department of Biological Chemistry, Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109, USA.