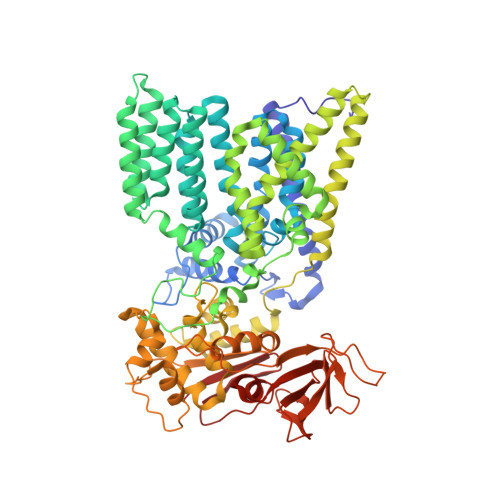
Molecular basis of lipid-linked oligosaccharide recognition and processing by bacterial oligosaccharyltransferase.
Napiorkowska, M., Boilevin, J., Sovdat, T., Darbre, T., Reymond, J.L., Aebi, M., Locher, K.P.(2017) Nat Struct Mol Biol 24: 1100-1106
- PubMed: 29058712
- DOI: https://doi.org/10.1038/nsmb.3491
- Primary Citation of Related Structures:
5OGL - PubMed Abstract:
Oligosaccharyltransferase (OST) is a membrane-integral enzyme that catalyzes the transfer of glycans from lipid-linked oligosaccharides (LLOs) onto asparagine side chains, the first step in protein N-glycosylation. Here, we report the X-ray structure of a single-subunit OST, PglB from Campylobacter lari, trapped in an intermediate state bound to an acceptor peptide and a synthetic LLO analog. The structure reveals the role of the external loop EL5, present in all OST enzymes, in substrate recognition. Whereas the N-terminal half of EL5 binds LLO, the C-terminal half interacts with the acceptor peptide. The glycan moiety of LLO must thread under EL5 to access the active site. Reducing EL5 mobility decreases the catalytic rate of OST when full-size heptasaccharide LLO is provided, but not for a monosaccharide-containing LLO analog. Our results define the chemistry of a ternary complex state, assign functional roles to conserved OST motifs, and provide opportunities for glycoengineering by rational design of PglB.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zurich, Zurich, Switzerland.