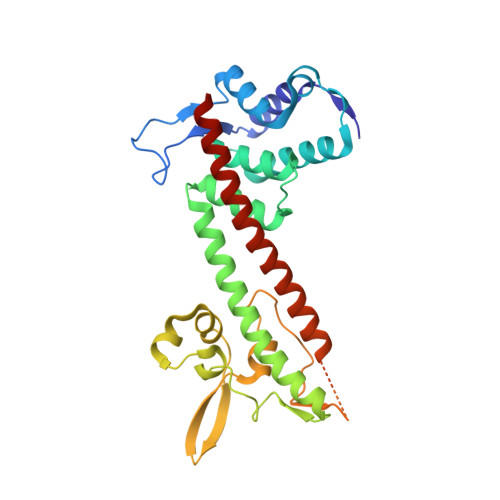
Structure of the Yersinia pestis tip protein LcrV refined to 1.65A resolution
Chaudhury, S., Battaile, K.P., Lovell, S., Plano, G.V., De Guzman, R.N.(2013) Acta Crystallogr Sect F Struct Biol Cryst Commun 69: 477-481
- PubMed: 23695558
- DOI: https://doi.org/10.1107/S1744309113008579
- Primary Citation of Related Structures:
4JBU - PubMed Abstract:
The human pathogen Yersinia pestis requires the assembly of the type III secretion system (T3SS) for virulence. The structural component of the T3SS contains an external needle and a tip complex, which is formed by LcrV in Y. pestis. The structure of an LcrV triple mutant (K40A/D41A/K42A) in a C273S background has previously been reported to 2.2 Å resolution. Here, the crystal structure of LcrV without the triple mutation in a C273S background is reported at a higher resolution of 1.65 Å. Overall the two structures are similar, but there are also notable differences, particularly near the site of the triple mutation. The refined structure revealed a slight shift in the backbone positions of residues Gly28-Asn43 and displayed electron density in the loop region consisting of residues Ile46-Val63, which was disordered in the original structure. In addition, the helical turn region spanning residues Tyr77-Gln95 adopts a different orientation.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA.