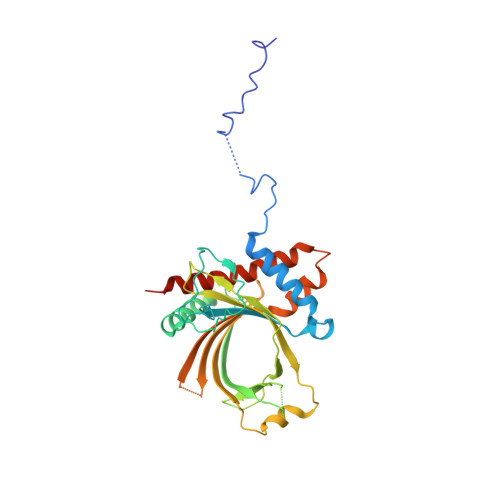
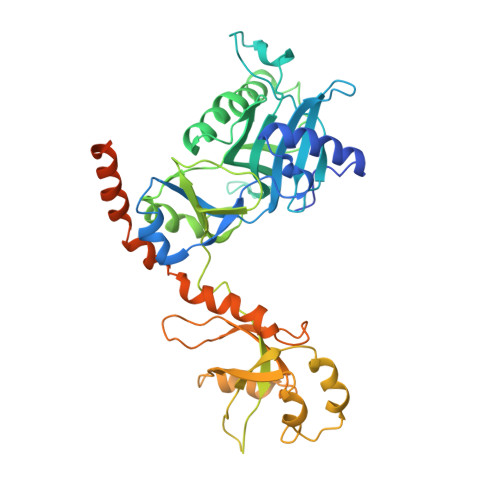
Structure of the Saccharomyces cerevisiae Cet1-Ceg1 mRNA Capping Apparatus.
Gu, M., Rajashankar, K.R., Lima, C.D.(2010) Structure 18: 216-227
- PubMed: 20159466
- DOI: https://doi.org/10.1016/j.str.2009.12.009
- Primary Citation of Related Structures:
3KYH - PubMed Abstract:
The 5' guanine-N7 cap is the first cotranscriptional modification of messenger RNA. In Saccharomyces cerevisiae, the first two steps in capping are catalyzed by the RNA triphosphatase Cet1 and RNA guanylyltransferase Ceg1, which form a complex that is directly recruited to phosphorylated RNA polymerase II (RNAP IIo), primarily via contacts between RNAP IIo and Ceg1. A 3.0 A crystal structure of Cet1-Ceg1 revealed a 176 kDa heterotetrameric complex composed of one Cet1 homodimer that associates with two Ceg1 molecules via interactions between the Ceg1 oligonucleotide binding domain and an extended Cet1 WAQKW amino acid motif. The WAQKW motif is followed by a flexible linker that would allow Ceg1 to achieve conformational changes required for capping while maintaining interactions with both Cet1 and RNAP IIo. The impact of mutations as assessed through genetic analysis in S. cerevisiae is consonant with contacts observed in the Cet1-Ceg1 structure.
Organizational Affiliation:
Structural Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.