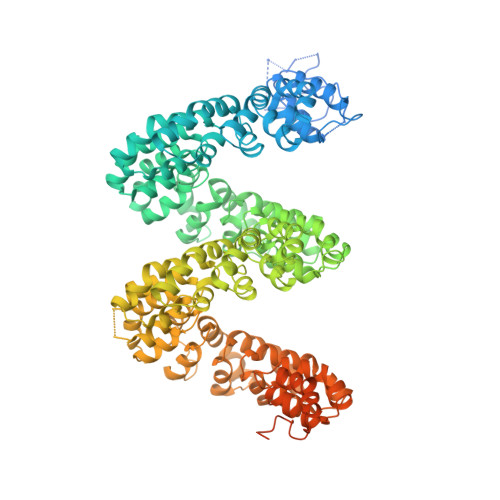
The Crystal Structure of TAL Effector PthXo1 Bound to Its DNA Target.
Mak, A.N., Bradley, P., Cernadas, R.A., Bogdanove, A.J., Stoddard, B.L.(2012) Science 335: 716-719
- PubMed: 22223736
- DOI: https://doi.org/10.1126/science.1216211
- Primary Citation of Related Structures:
3UGM - PubMed Abstract:
DNA recognition by TAL effectors is mediated by tandem repeats, each 33 to 35 residues in length, that specify nucleotides via unique repeat-variable diresidues (RVDs). The crystal structure of PthXo1 bound to its DNA target was determined by high-throughput computational structure prediction and validated by heavy-atom derivatization. Each repeat forms a left-handed, two-helix bundle that presents an RVD-containing loop to the DNA. The repeats self-associate to form a right-handed superhelix wrapped around the DNA major groove. The first RVD residue forms a stabilizing contact with the protein backbone, while the second makes a base-specific contact to the DNA sense strand. Two degenerate amino-terminal repeats also interact with the DNA. Containing several RVDs and noncanonical associations, the structure illustrates the basis of TAL effector-DNA recognition.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, A3-025 Seattle, WA 98019, USA.