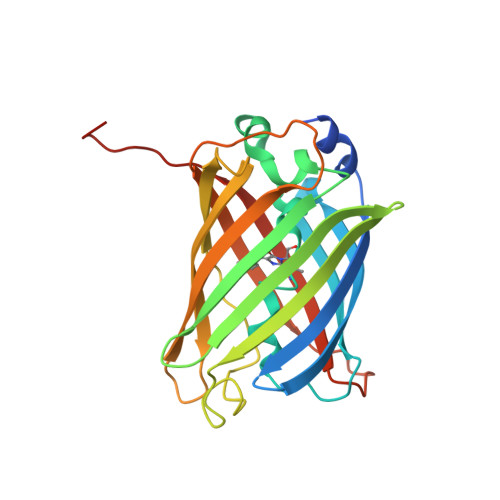
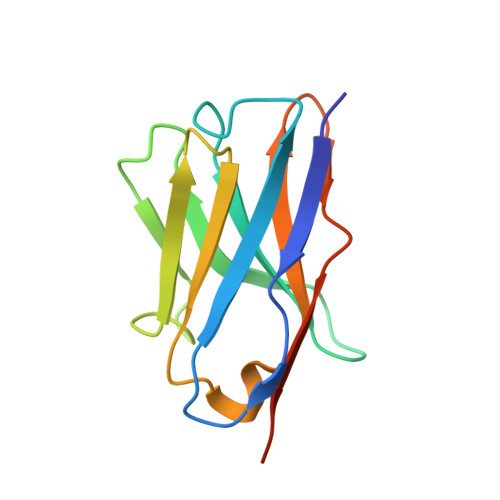
Structural and thermodynamic analysis of the GFP:GFP-nanobody complex.
Kubala, M.H., Kovtun, O., Alexandrov, K., Collins, B.M.(2010) Protein Sci 19: 2389-2401
- PubMed: 20945358
- DOI: https://doi.org/10.1002/pro.519
- Primary Citation of Related Structures:
3OGO - PubMed Abstract:
The green fluorescent protein (GFP)-nanobody is a single-chain VHH antibody domain developed with specific binding activity against GFP and is emerging as a powerful tool for isolation and cellular engineering of fluorescent protein fusions in many different fields of biological research. Using X-ray crystallography and isothermal titration calorimetry, we determine the molecular details of GFP:GFP-nanobody complex formation and explain the basis of high affinity and at the same time high specificity of protein binding. Although the GFP-nanobody can also bind YFP, it cannot bind the closely related CFP or other fluorescent proteins from the mFruit series. CFP differs from GFP only within the central chromophore and at one surface amino acid position, which lies in the binding interface. Using this information, we have engineered a CFP variant (I146N) that is also able to bind the GFP-nanobody with high affinity, thus extending the toolbox of genetically encoded fluorescent probes that can be isolated using the GFP-nanobody.
Organizational Affiliation:
Institute for Molecular Bioscience, University of Queensland, St. Lucia, Queensland 4072, Australia.