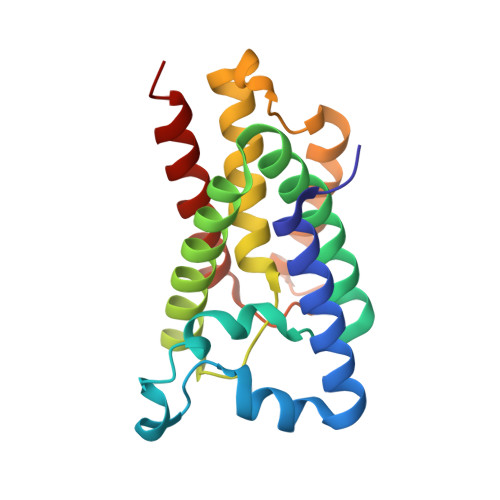
Crystal structure of a rhomboid family intramembrane protease.
Wang, Y., Zhang, Y., Ha, Y.(2006) Nature 444: 179-180
- PubMed: 17051161
- DOI: https://doi.org/10.1038/nature05255
- Primary Citation of Related Structures:
2IC8 - PubMed Abstract:
Escherichia coli GlpG is an integral membrane protein that belongs to the widespread rhomboid protease family. Rhomboid proteases, like site-2 protease (S2P) and gamma-secretase, are unique in that they cleave the transmembrane domain of other membrane proteins. Here we describe the 2.1 A resolution crystal structure of the GlpG core domain. This structure contains six transmembrane segments. Residues previously shown to be involved in catalysis, including a Ser-His dyad, and several water molecules are found at the protein interior at a depth below the membrane surface. This putative active site is accessible by substrate through a large 'V-shaped' opening that faces laterally towards the lipid, but is blocked by a half-submerged loop structure. These observations indicate that, in intramembrane proteolysis, the scission of peptide bonds takes place within the hydrophobic environment of the membrane bilayer. The crystal structure also suggests a gating mechanism for GlpG that controls substrate access to its hydrophilic active site.
Organizational Affiliation:
Department of Pharmacology, Yale School of Medicine, 333 Cedar Street, New Haven, Connecticut 06520, USA.