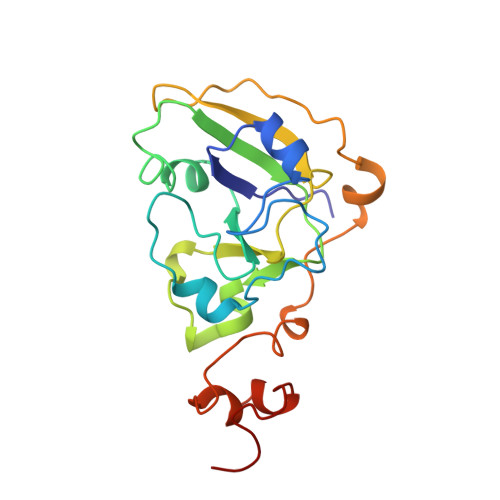
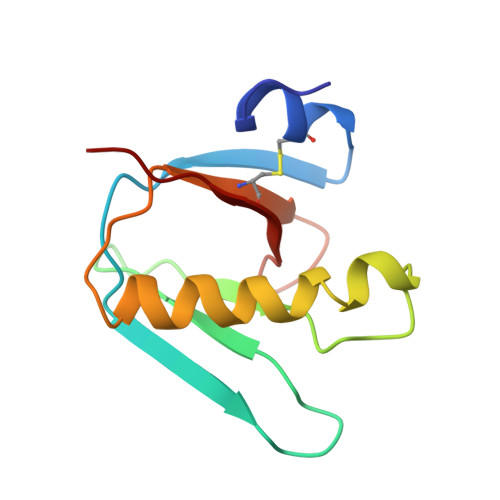
The three-dimensional crystal structure of cholera toxin.
Zhang, R.G., Scott, D.L., Westbrook, M.L., Nance, S., Spangler, B.D., Shipley, G.G., Westbrook, E.M.(1995) J Mol Biol 251: 563-573
- PubMed: 7658473
- DOI: https://doi.org/10.1006/jmbi.1995.0456
- Primary Citation of Related Structures:
1XTC - PubMed Abstract:
The clinical manifestations of cholera are largely attributable to the actions of a secreted hexameric AB5 enterotoxin (choleragen). We have independently solved and refined the three-dimensional structure of choleragen at 2.5 A resolution. The structure of the crystalline toxin closely resembles that described for the heat-labile enterotoxin from Escherichia coli (LT) with which it shares 80% sequence homology. In both cases, the wedge-shaped A subunit is loosely held high above the plane of the pentameric B subunits by the tethering A2 chain. The most striking difference between the two toxins occurs at the carboxyl terminus of the A2 chain. Whereas the last 14 residues of the A2 chain of LT threading through the central pore of the B5 assembly form an extended chain with a terminal loop, the A2 chain of choleragen remains a nearly continuous alpha-helix throughout its length. The four carboxyl-terminal residues of the A2 chain (KDEL sequence), disordered in the crystal structure of LT, are clearly visible in choleragen's electron-density map. In the accompanying article we describe the three-dimensional structure of the isolated B pentamer of cholera toxin (choleragenoid). Comparison of the crystalline coordinates of choleragen, choleragenoid, and LT provides a solid three-dimensional foundation for further experimental investigation. These structures, along with those of related toxins from Shigella dysenteria and Bordetella pertussis, offer a first step towards the rational design of new vaccines and anti-microbial agents.
Organizational Affiliation:
Center for Mechanistic Biology and Biotechnology, Argonne National Laboratory, IL 60439, USA.