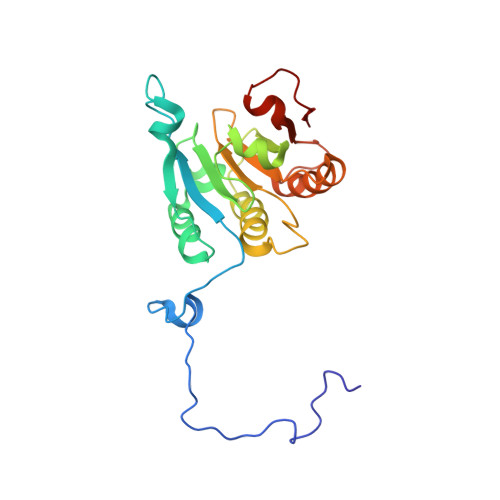
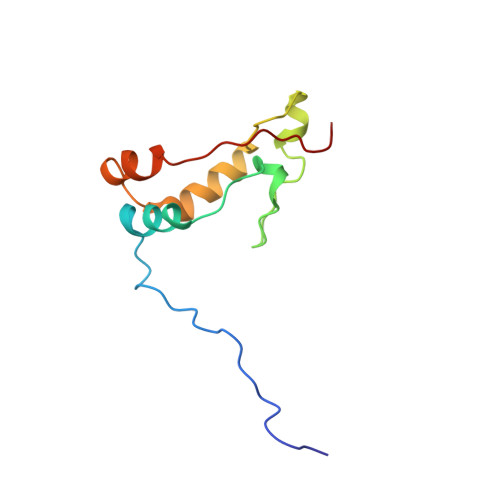
Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Gross, J.D., Moerke, N.J., von der Haar, T., Lugovskoy, A.A., Sachs, A.B., McCarthy, J.E., Wagner, G.(2003) Cell 115: 739-750
- PubMed: 14675538
- DOI: https://doi.org/10.1016/s0092-8674(03)00975-9
- Primary Citation of Related Structures:
1RF8 - PubMed Abstract:
The eukaryotic initiation factor 4G (eIF4G) is the core of a multicomponent switch controlling gene expression at the level of translation initiation. It interacts with the small ribosomal subunit interacting protein, eIF3, and the eIF4E/cap-mRNA complex in order to load the ribosome onto mRNA during cap-dependent translation. We describe the solution structure of the complex between yeast eIF4E/cap and eIF4G (393-490). Binding triggers a coupled folding transition of eIF4G (393-490) and the eIF4E N terminus resulting in a molecular bracelet whereby eIF4G (393-490) forms a right-handed helical ring that wraps around the N terminus of eIF4E. Cofolding allosterically enhances association of eIF4E with the cap and is required for maintenance of optimal growth and polysome distributions in vivo. Our data explain how mRNA, eIF4E, and eIF4G exists as a stable mRNP that may facilitate multiple rounds of ribosomal loading during translation initiation, a key determinant in the overall rate of protein synthesis.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.