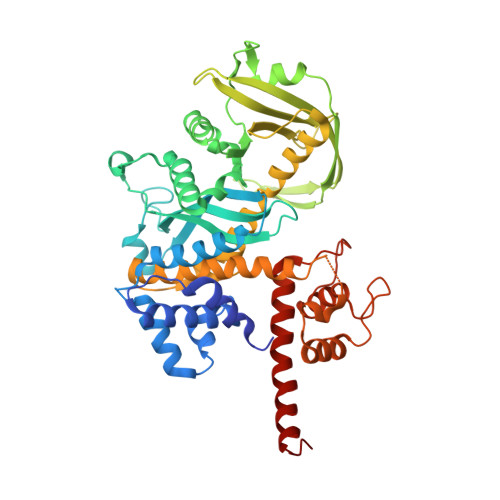
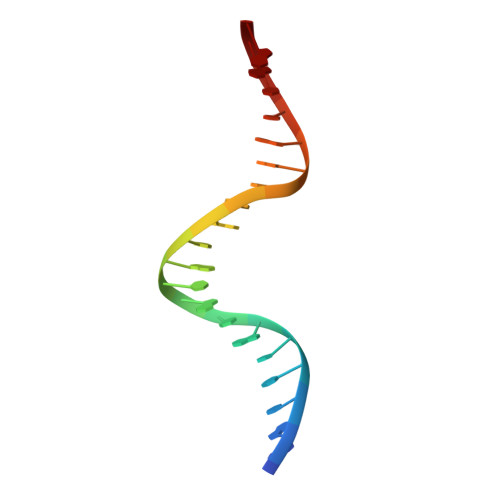
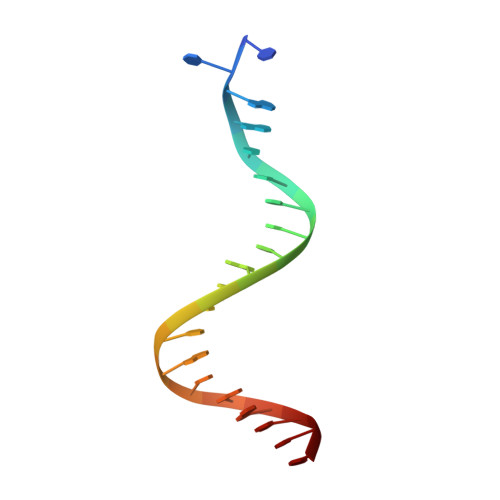
Three-dimensional structure of the Tn5 synaptic complex transposition intermediate.
Davies, D.R., Goryshin, I.Y., Reznikoff, W.S., Rayment, I.(2000) Science 289: 77-85
- PubMed: 10884228
- DOI: https://doi.org/10.1126/science.289.5476.77
- Primary Citation of Related Structures:
1MUH - PubMed Abstract:
Genomic evolution has been profoundly influenced by DNA transposition, a process whereby defined DNA segments move freely about the genome. Transposition is mediated by transposases, and similar events are catalyzed by retroviral integrases such as human immunodeficiency virus-1 (HIV-1) integrase. Understanding how these proteins interact with DNA is central to understanding the molecular basis of transposition. We report the three-dimensional structure of prokaryotic Tn5 transposase complexed with Tn5 transposon end DNA determined to 2.3 angstrom resolution. The molecular assembly is dimeric, where each double-stranded DNA molecule is bound by both protein subunits, orienting the transposon ends into the active sites. This structure provides a molecular framework for understanding many aspects of transposition, including the binding of transposon end DNA by one subunit and cleavage by a second, cleavage of two strands of DNA by a single active site via a hairpin intermediate, and strand transfer into target DNA.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin, Madison, WI 53706, USA.