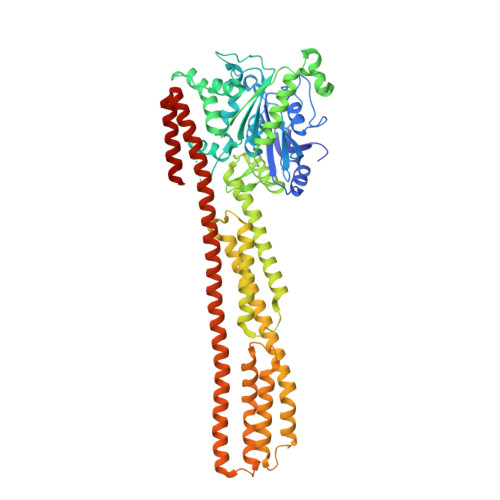
Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism.
Prakash, B., Renault, L., Praefcke, G.J., Herrmann, C., Wittinghofer, A.(2000) EMBO J 19: 4555-4564
- PubMed: 10970849
- DOI: https://doi.org/10.1093/emboj/19.17.4555
- Primary Citation of Related Structures:
1F5N - PubMed Abstract:
The interferon-gamma-induced guanylate-binding protein 1 (GBP1) belongs to a special class of large GTP- binding proteins of 60-100 kDa with unique characteristics. Here we present the structure of human GBP1 in complex with the non-hydrolysable GTP analogue GppNHp. Basic features of guanine nucleotide binding, such as the P-loop orientation and the Mg(2+) co-ordination, are analogous to those of Ras-related and heterotrimeric GTP-binding proteins. However, the glycosidic bond and thus the orientation of the guanine base and its interaction with the protein are very different. Furthermore, two unique regions around the base and the phosphate-binding areas, the guanine and the phosphate caps, respectively, give the nucleotide-binding site a unique appearance not found in the canonical GTP-binding proteins. The phosphate cap, which constitutes the region analogous to switch I, completely shields the phosphate-binding site from solvent such that a potential GTPase-activating protein cannot approach. This has consequences for the GTPase mechanism of hGBP1 and possibly of other large GTP-binding proteins.
Organizational Affiliation:
Max-Planck-Institut für molekulare Physiologie, Otto-Hahn-Strabetae 11, D-44227 Dortmund, Germany.